Stochastic Gradient Descent Framework¶
Stochasitc gradient descent is a method to optimize an objective function has certain properties. It is similar to gradient descent but different in data loaded. Gradient Descent uses whole set of data, while SGD randomly choose a fraction of data from the whole set.
Since random access data in DDR will have poor effiency on DDR bandwidth. We implement a “Drop or Jump” table sampler. If choose “drop”, the sampler will continuously read data from the table, and drop a part of them. This will lead to continuously burst read of data. This is better when fraction is not too small. If choose “jump”, the sampler will read a continuous bucket data, jump a few buckets and read the next bucket. This will lead to burst read of data of certain length and interupted by jump. This is better when fraction is relatively small. In such way, we could have better DDR access efficiency.
Each iteration, SGD framework will compute gradient of currrent weigth (and intercept if needed). Then SGD will update weight according to gradient. Linear Least Sqaure Regression, LASSO Regression and Ridge Regression training share the same gradient calculation process. There’re 3 different way to update: Simple update, L1 update and L2 update. They’ll have different traing result and various desired characteristics.
Linear Least Sqaure Regression Training¶
Linear Least Square Regression uses simple update:
LASSO Regression Training¶
LASSO Regression uses L1 update:
Ridge Regression Training¶
Ridge Regression uses L2 Update:
Implementation (Training)¶
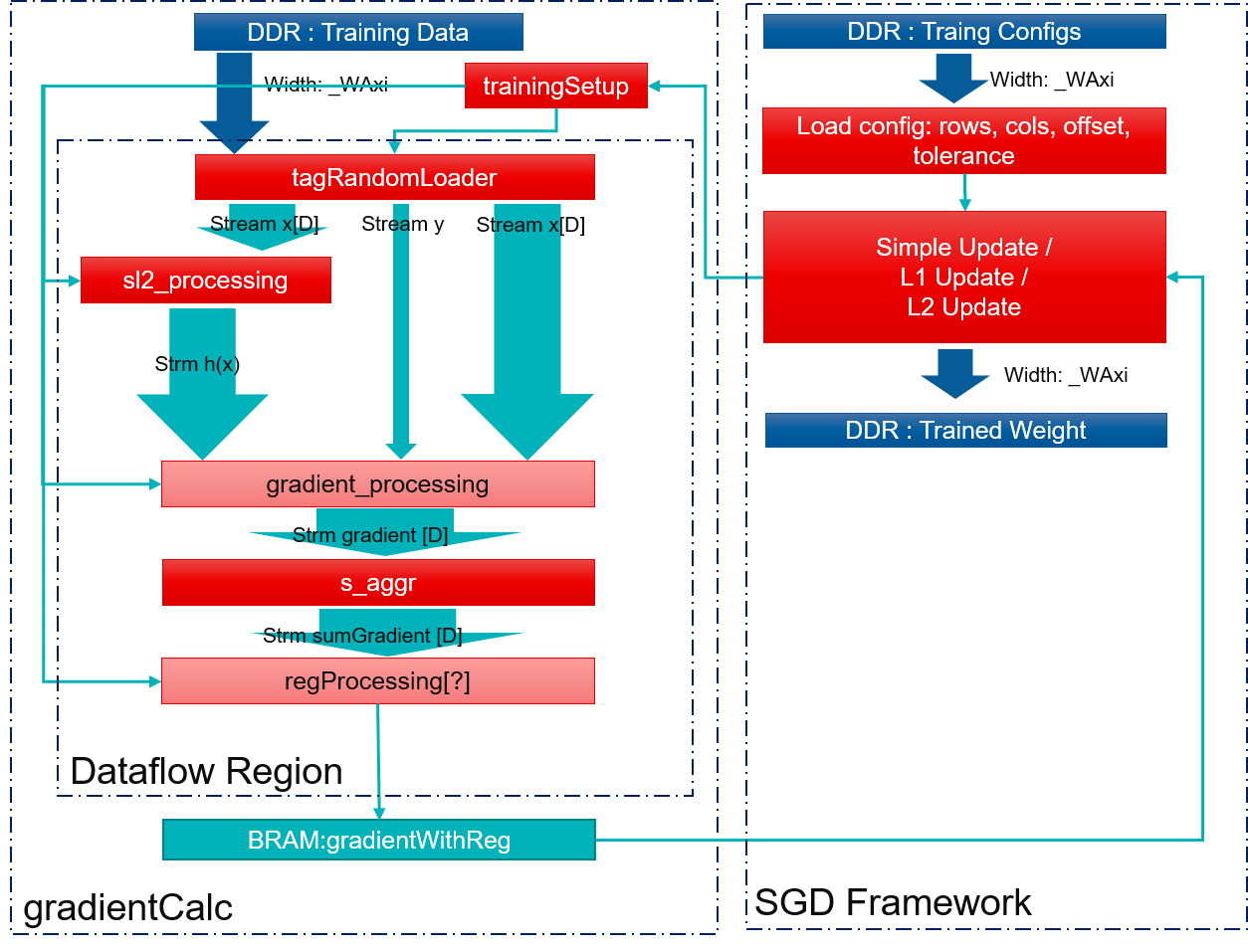
SGD Framework is basically two parts: gradient calculation and weight update. Gradient calculation load data from DDR and calculate gradient of preset weight and is a dataflow function. Weight update calculate next weight based on gradient and chosen method and is executed after gradient calculation. Block diagram is shown as below.
The correctness of Linear Regression/LASSO Regression/Ridge Regression Training using SGD framework is verified by comparing results with Spark mllib. The results are identical.